Published Pictures for 26919 Rab11-GTP mAb
Rab11a-GTP measurement assay
Rab11a activity was measured in all strains using a conformation specific antibody that specifically recognizes Rab11-GTP (NewEast Biosciences catalogue number 26919). For in vitro precipitation of Rab11a-GTP, 1 μg bacterially expressed GST Rab was incubated with 20mM EDTA and either 100 μM GTPγS or 1 mM GDP for 30 minutes at 30°C. GTP/GDP loading was stopped by placing the tubes on ice and adding 60 mM MgCl2. Reaction volume was adjusted to 1 ml with lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% Triton X-100) before incubating with 1 μl of anti Rab11-GTP antibody and 20 μl of protein A/G agarose (50% slurry) for 1 hour at 4°C with gentle agitation. Beads were washed three times in lysis buffer before elution of bound protein by boiling in 20 μl of 2X SDS sample buffer. Proteins were separated by SDS-PAGE and immunoblotted for GST using an anti-GST antibody (Abcam catalogue number ab6613). For in vivo precipitation of rab11a-GTP, 1×107 cells expressing Rab11a-RFP were lysed in 1ml lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% Triton X-100). Lysate was then incubated with 1 μl of anti Rab11-GTP antibody and 20 μl of protein A/G agarose (50% slurry) for 1 hour at 4°C with gentle agitation. Beads were washed three times in lysis buffer before elution of bound protein by boiling in 20 μl of 2X SDS sample buffer. Proteins from input, bound and not bound fractions were separated by SDS-PAGE and immunoblotted for RFP using an anti-RFP antibody (Chromotek catalogue number 3F5). Anti-actin antibody (Santa Cruz Biotechnology catalogue number sc-47778) was used as a loading control for each sample. The ratio of bound Rab11a-RFP (Rab11a-GTP) and unbound Rab11a-RFP (Rab11a-GDP) was calculated for each clone after normalizing against the actin loading control. Densitometry analyses of western blots was performed in imageJ. All blots analysed were in the linear range.
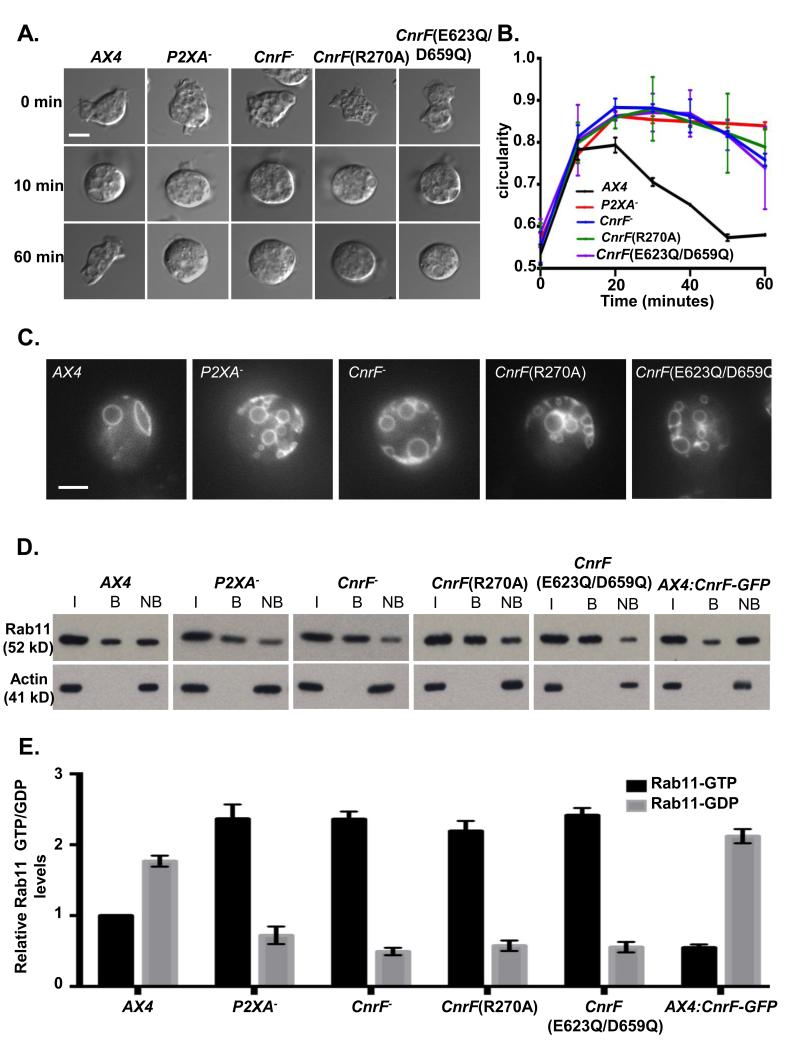
cnrF is required for normal osmoregulation and regulation of Rab11 activity
D and E. Immunoprecipitation of Rab11a-GTP from wild-type, P2XA− cnrF−, cnrF(R270A), cnrF(E623Q/D659Q) and CnrF-GFP overexpressing cells expressing Rab11a-RFP (I = input, B = bound, NB = not bound). The level of Rab11a-RFP expression (input) is comparable between all strains (>1.3 fold difference between highest and lowest). E. Quantification of GTP and GDP bound Rab11a revealed that all mutant strains exhibit a significant (paired T test p<0.001) 2-2.5 fold increase in the levels of Rab11a-GTP compared to wild type cells. Furthermore CnrF-GFP overexpressing cells exhibit a reciprocal 2 fold decrease in GTP bound Rab11a compare to wild type (paired T test p<0.001). Error bars represent s.e.m. from n=3 three independent experiments.
Rab11 activity assay
Rab11 activity was determined using a Rab11 activity assay kit (NewEast Biosciences, King of Prussia, PA) according to manufacturer recommended protocol. HSC lysates containing equal amounts of total proteins were incubated with a mouse monoclonal antibody recognizing GTP bound Rab11 specifically. The bound active Rab11 was pulled down by protein A/G agarose and detected by a rabbit polyclonal anti-Rab11 antibody.
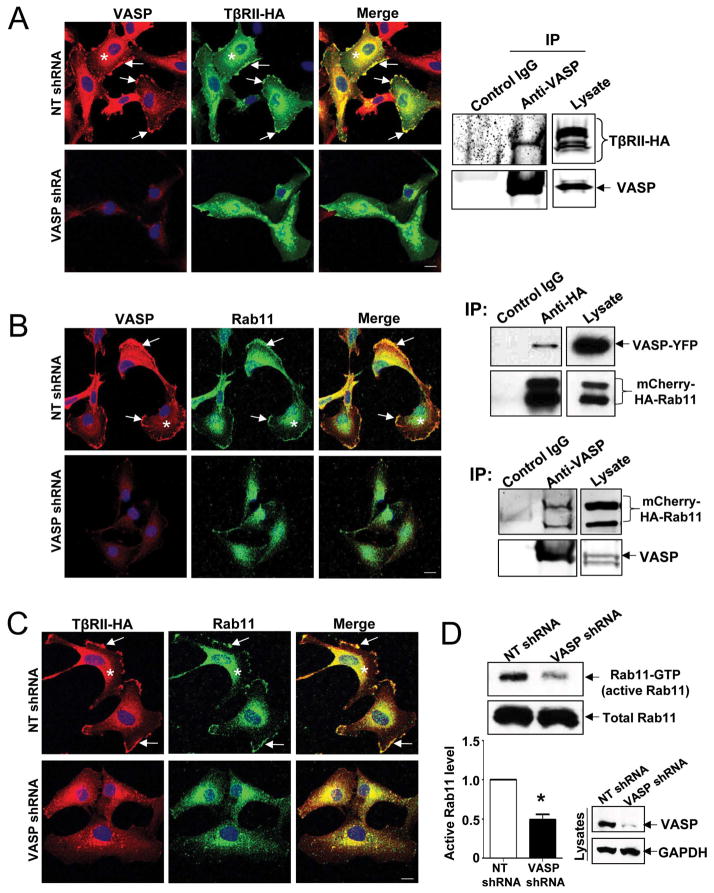
VASP is required for targeting of VASP/TβRII/Rab11 complexes to the plasma membrane and VASP knockdown inhibits Rab11 activity
…(D) HSCs with or without VASP knockdown were subjected to Rab11 activity assays. VASP knockdown significantly reduced amounts of GTP bound Rab11 (active form of Rab11) of HSCs. *p < 0.05 by t test. n=3.
Rab11a-GTP pull-down assay
Rab11a pull-down assay was performed using Rab11 activation assay kit (Neweast Bioscience) according to the manufacturer’s instructions (17). Cells were washed once with ice-cold PBS and lysed with cell lysis buffer. Lysates were centrifuged at 13,000 × g for 10 min, and the supernatant was incubated with 1.0 μg anti-active Rab11 Ab coupled with protein A/G agarose. The reaction mixture was gently rocked for 1 h at 4°C. The beads were then collected by centrifugation at 5000 × g, incubated at 4°C for 1 min, and washed twice with lysis buffer. Beads were finally resuspended in Laemmli sample buffer (30 μl) and boiled for immunoblotting with anti-Rab11a Ab. The total level of Rab11a was measured by Western blot analysis of cell lysates.
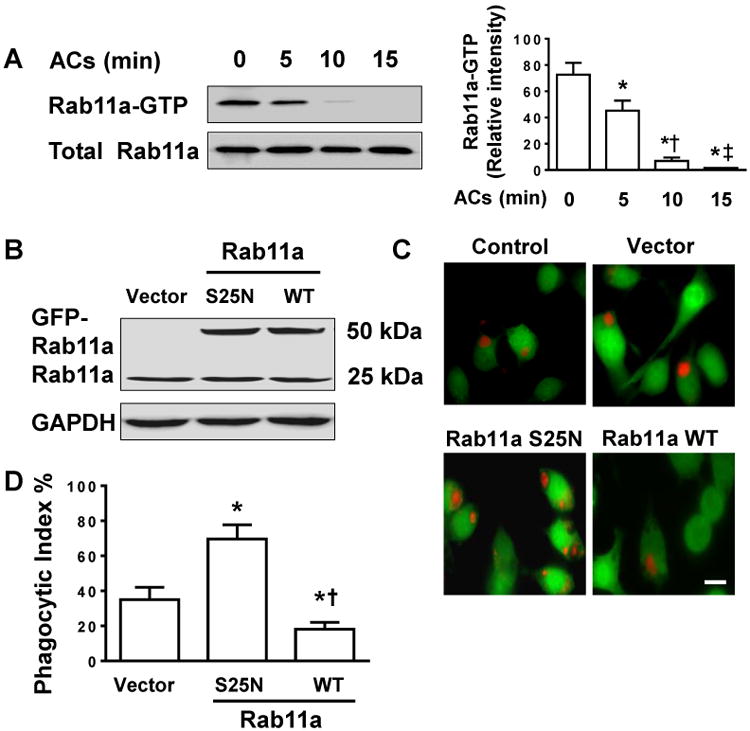
Rab11a Inactivation promotes clearance of apoptotic PMNs
(A) Time-dependent Rab11a inactivation in BMDMs in response to apoptotic PMNs (ACs) feeding. BMDMs were co-cultured with apoptotic PMNs at 1:10 ratio for ∼15 min. Lysates from macrophages were analyzed by Rab11a Activation Assay Kit. Left, pull-down experiment showing the content of Rab11-GTP in BMDMs stimulated with ACs; Right, quantification of three independent experiments is provided. (B) Endogenous and exogenous expression of Rab11a protein in BMDMs analyzed by Western blots. BMDMs were transfected with vector, dominant negative (S25N), or wild type (WT) Rab11a cDNA. (C) Representative fluorescent imaging of phagocytosis of ACs. The transfected BMDMs were labeled with CellTracker™ Green. After incubation of macrophages with CellTracker™ Red-labelled apoptotic PMNs for 2 h, macrophage efferocytosis was visualized using fluorescence microscopy. Scale bar, 10 μm. (D) Phagocytic index. Data were obtained from three independent cultures, each performed in triplicate, and expressed as mean ± SEM. The P value was calculated by one-way ANOVA followed by Student’s t-test. *P<0.05 vs. control (A) or vector group (D); †P <0.05 vs. 5 min group (A) or S25N group (D); ‡ P <0.05 vs. 10 min group (A).
Immunoprecipitation
HEK293 cells were transiently transfected with the indicated constructs and were maintained as described above for 48 hours. The cells were then washed with ice-cold PBS and harvested in 300 µl of lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 0.5% deoxycholate, 0.1% SDS, 10 mM Na4P2O7, 1% IGEPAL, and 5 mM EDTA or 1 mM CaCl2 depending on the antibody used for the assay) or ubiquitylation lysis buffer for ubiquitylation experiments (50 mM HEPES, pH 7.5, 250 mM NaCl, 2 mM EDTA or 1 mM CaCl2, 0.5% IGEPAL, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4, 10% glycerol and 10 mM N-ethylmaleimide). Both buffers were supplemented with protease inhibitors (9 nM pepstatin, 9 nM antipain, 10 nM leupeptin and 10 nM chymostatin) (Sigma Aldrich). Immunoprecipitations were then carried out as we described previously (Lachance et al., 2011; Parent et al., 2010).
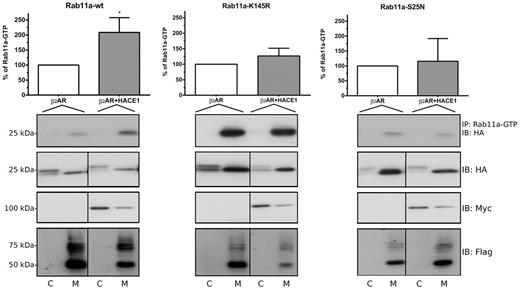
Ubiquitylation of Rab11a on Lys145 by β2AR-HACE1 is involved in its activation. HEK293 cells were transfected with HA-Rab11a, HA-Rab11a-K145R or HA-Rab11a-S25N in combination with FLAG-β2AR alone or with HACE1-Myc. Cells were subjected to fractionation 48 hours after transfection. Cytosolic (C) or membrane (M) fractions were immunoprecipitated with an antibody specifically recognizing activated Rab11-GTP. Immunoprecipitates and samples from the cytoplasmic and membrane fractions were analyzed by western blot using the indicated antibodies. The blots shown are representative of four separate experiments. Densitometry analyses of the quantity of Rab11-GTP that were immunoprecipitated from the membrane fractions were performed with ImageJ software. Densitometry value measured for the β2AR condition was set at 100%. Results are the means ± s.e.m. of four independent experiments. The statistical significance was determined using an unpaired Student’s t-test. *P<0.05.
Rab11a activity assay
Rab11a activation was determined by using Rab11 activation assay kit (NB-83201) purchased from NewEast Biosciences (PA, USA). The procedure was performed as previously described (Yan et al.2016). Briefly, the mixture of 1 mg cell lysate and 1 µL antiactive Rab11 antibodies were resuspended in protein-A/G agarose beads slurry (20 µL). After incubation for 1 h at 4 °C, the beads were washed 3 times with 0.5 mL assay buffer, and then resuspended in SDS-PAGE sample buffer for further Western blot analysis.
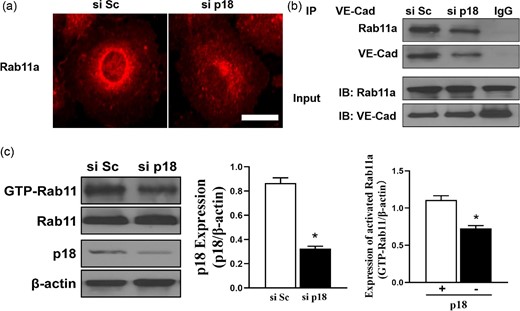
p18 plays an important role in VE-cadherin recycling by affecting Rab11a. (a) Immunofluorescence analysis of Rab11a-positive recycling endosomes in p18 knockdown hPMVECs. (b) Association of Rab11a and VE-cadherin in hPMVECs transfected with p18 siRNA (si p18) or scrambled (si Sc). Co-immunoprecipitation were performed and the association between Rab11a and VE-cadherin were analyzed. Input: Total cell lysates. (c) p18 induced Rab11a activation. Left: Western blotting analysis was performed in hPMVECs transfected with scrambled (si Sc) or p18 siRNA (si p18). Middle: Quantitative analysis of p18 expression after si p18 transfection; n = 3. *P < .05 vs si Sc group. Right: Quantitative analysis of the activation level of rab11a after p18 knockdown; n = 3. *P < .05 vs si Sc group.
Western blotting
Protein lysates were boiled with Laemmli buffer (Bio-Rad), separated on SDS polyacrylamide gels and transferred onto nitrocellulose membranes. Blots were blocked with 5% nonfat dry milk then probed with mouse monoclonal anti-BKα (1:500 dilution, Neuromab, UC Davis), rabbit polyclonal anti-BKβ1 (1:500, Abcam), rabbit monoclonal anti-rab11A (1:500, Cell Signaling), rabbit polyclonal anti-phosphoserine (1:500, Millipore) or mouse monoclonal anti-active rab11 (1:500, NewEast Biosciences) antibodies overnight at 4°C. After incubating with horseradish peroxidase-conjugated secondary antibodies and a chemiluminescent detection kit, the blots were imaged with a Kodak In Vivo F Pro Imaging System. Band intensity was quantified using Quantity One software (Bio-Rad).
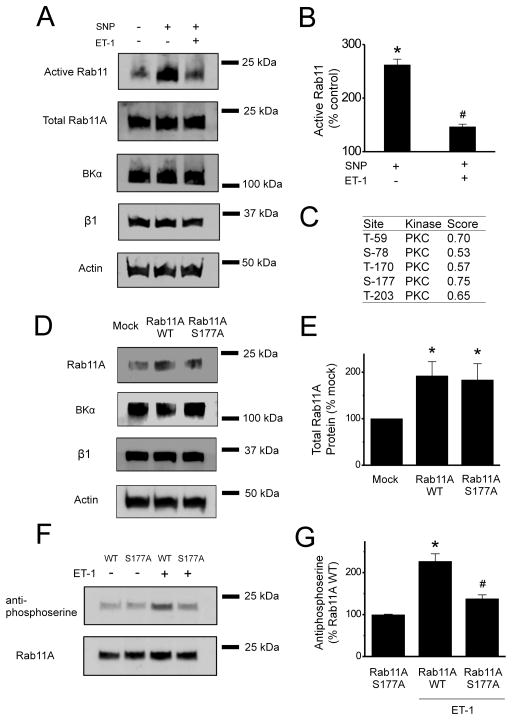
ET-1 and NO modulate Rab11 activity and ET-1 stimulates phosphorylation of Rab11A at serine 177
(A) Representative Western Blots of active Rab11 and total Rab11A, BKα, β1 and actin in control arteries and arteries treated with SNP (10 μmol/L, 10 min) or SNP + ET-1 (10 nmol/L, 1h). (B) Mean data of active Rab11 compared to control (n=6). *p<0.05 vs. control, # p<0.05 vs SNP. (C) Potential PKC phosphorylation sites in Rab11A, of which S177 has the highest probability. (D) Representative Western Blots probed for total Rab11A, BKα, β1 or actin protein in arteries transfected with either an empty vector (mock) or a vector encoding Rab11A WT or Rab11A S177A. (E) Mean data of Rab11A total protein compared to mock (n=6). *p <0.05 vs. mock group. (F) Representative Western Blots probed with anti-phosphoserine or Rab11A antibodies after immunoprecipitation of Rab11A. (G) Mean data of phosphoserine band intensity compared to untreated Rab11A WT (n=6). *p <0.05 vs. Rab11A WT without ET-1 treatment, # p <0.05 vs. Rab11A WT with ET-1 treatment.

Rab-GTP activity analysis
Activation of Rab5, Rab7, and Rab11 were analysed with Rab5, Rab7, and Rab11 Activation Assay Kits (NewEast Biosciences, King of Prussia, PA, USA). Briefly, 500 μg total protein lysed with 1× lysis buffer per sample were used in a Rab-GTP pull-down assay. Each sample was combined with anti-Rab-GTP monoclonal antibody and incubated at 4 °C overnight. Protein A/G agarose beads were used to capture the antibodies. After 1 h incubation at 4 °C, the beads were boiled in 2× loading buffer for 5 min. The supernatant was used for Western blot to detect Rab-GTP activity using Rab polyclonal antibody.
DPBA induces EGFR degradation in a lysosome-dependent manner.
…... h DPBA induced EGFR perinuclear accumulation. A549 and H1975 were treated with DPBA (6 μM) for 6 h. EGFR sublocalisation was detected by immunofluorescence (magnification, ×200; scale bar, 50 μm). i DPBA induced surface EGFR endocytosis. A549 and H1975 were treated with DPBA (6 μM) for 3 and 6 h. Surface and intracellular EGFR were measured by biotinylation assay. TfR was the loading control for surface protein. j DPBA induced EGFR trafficking through endo-lysosome route. A549 was treated with DPBA (6 μM) for 3 and 6 h. Colocalisation between EGFR and LAMP1, Rab5, Rab7, or Rab11 was detected by immunofluorescence (magnification, ×630; scale bar, 10 μm). k DPBA activated Rab5 and Rab7, but not Rab11. A549 was treated with DPBA (6 μM) for 3, 6, and 12 h. Rab-GTP expression was measured with a Rab Activation Assay Kit

Rab-GTP activity analysis
Activation of Rab5, Rab7, and Rab11 were analysed with Rab5, Rab7, and Rab11 Activation Assay Kits (NewEast Biosciences, King of Prussia, PA, USA). Briefly, 500 μg total protein lysed with 1× lysis buffer per sample were used in a Rab-GTP pull-down assay. Each sample was combined with anti-Rab-GTP monoclonal antibody and incubated at 4 °C overnight. Protein A/G agarose beads were used to capture the antibodies. After 1 h incubation at 4 °C, the beads were boiled in 2× loading buffer for 5 min. The supernatant was used for Western blot to detect Rab-GTP activity using Rab polyclonal antibody.
DPBA induces EGFR degradation in a lysosome-dependent manner.
…... h DPBA induced EGFR perinuclear accumulation. A549 and H1975 were treated with DPBA (6 μM) for 6 h. EGFR sublocalisation was detected by immunofluorescence (magnification, ×200; scale bar, 50 μm). i DPBA induced surface EGFR endocytosis. A549 and H1975 were treated with DPBA (6 μM) for 3 and 6 h. Surface and intracellular EGFR were measured by biotinylation assay. TfR was the loading control for surface protein. j DPBA induced EGFR trafficking through endo-lysosome route. A549 was treated with DPBA (6 μM) for 3 and 6 h. Colocalisation between EGFR and LAMP1, Rab5, Rab7, or Rab11 was detected by immunofluorescence (magnification, ×630; scale bar, 10 μm). k DPBA activated Rab5 and Rab7, but not Rab11. A549 was treated with DPBA (6 μM) for 3, 6, and 12 h. Rab-GTP expression was measured with a Rab Activation Assay Kit



